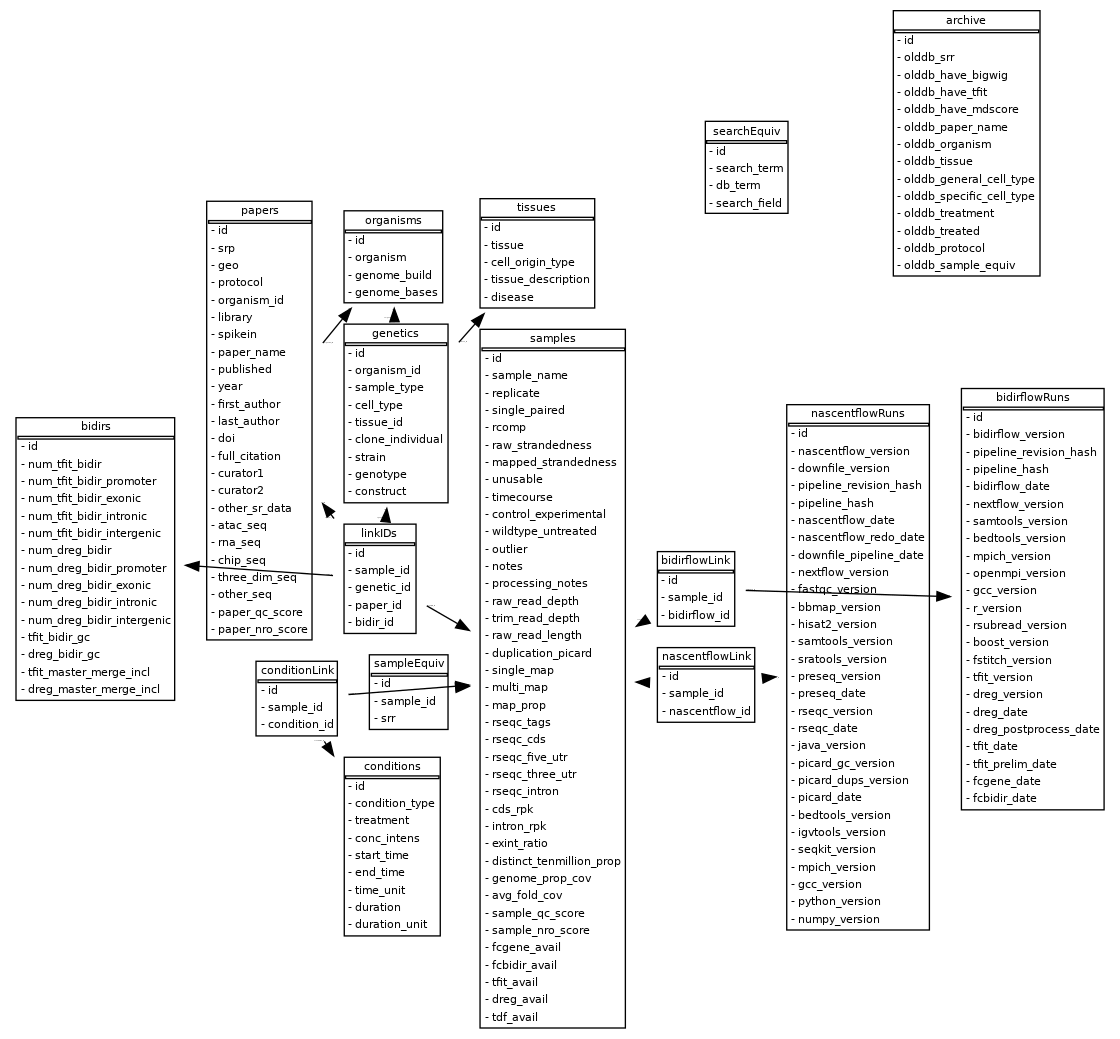
Database schema diagram
Database field types (alphabetical by field)
| Field name | Field type |
| atac_seq | Boolean |
| avg_fold_cov | Float |
| bbmap_version | Character |
| bedtools_version | Character |
| bedtools_version | Character |
| bidirflow_date | Date |
| bidirflow_version | Character |
| boost_version | Character |
| cds_rpk | Float |
| cell_origin_type | Character |
| cell_type | Character |
| chip_seq | Boolean |
| clone_individual | Character |
| conc_intens | Character |
| condition_type | Character |
| construct | Character |
| control_experimental | Character |
| curator1 | Character |
| curator2 | Character |
| db_term | Character |
| disease | Boolean |
| distinct_tenmillion_prop | Float |
| doi | Character |
| downfile_pipeline_date | Date |
| downfile_version | Character |
| dreg_avail | Boolean |
| dreg_bidir_gc | Float |
| dreg_date | Date |
| dreg_master_merge_incl | Boolean |
| dreg_postprocess_date | Date |
| dreg_version | Character |
| duplication_picard | Float |
| duration | Integer |
| duration_unit | Character |
| end_time | Integer |
| exint_ratio | Float |
| fastqc_version | Character |
| fcbidir_avail | Boolean |
| fcbidir_date | Date |
| fcgene_avail | Boolean |
| fcgene_date | Date |
| first_author | Character |
| fstitch_version | Character |
| full_citation | Character |
| gcc_version | Character |
| gcc_version | Character |
| genome_bases | Big Integer |
| genome_build | Character |
| genome_prop_cov | Float |
| genotype | Character |
| geo | Character |
| hisat2_version | Character |
| igvtools_version | Character |
| intron_rpk | Float |
| Field name | Field type |
| java_version | Character |
| last_author | Character |
| library | Character |
| map_prop | Float |
| mapped_strandedness | Character |
| mpich_version | Character |
| mpich_version | Character |
| multi_map | Integer |
| nascentflow_date | Date |
| nascentflow_redo_date | Date |
| nascentflow_version | Character |
| nextflow_version | Character |
| nextflow_version | Character |
| notes | Character |
| num_dreg_bidir | Integer |
| num_dreg_bidir_exonic | Integer |
| num_dreg_bidir_intergenic | Integer |
| num_dreg_bidir_intronic | Integer |
| num_dreg_bidir_promoter | Integer |
| num_tfit_bidir | Integer |
| num_tfit_bidir_exonic | Integer |
| num_tfit_bidir_intergenic | Integer |
| num_tfit_bidir_intronic | Integer |
| num_tfit_bidir_promoter | Integer |
| numpy_version | Character |
| olddb_general_cell_type | Character |
| olddb_have_bigwig | Boolean |
| olddb_have_mdscore | Boolean |
| olddb_have_tfit | Boolean |
| olddb_organism | Character |
| olddb_paper_name | Character |
| olddb_protocol | Character |
| olddb_sample_equiv | Character |
| olddb_specific_cell_type | Character |
| olddb_srr | Character |
| olddb_tissue | Character |
| olddb_treated | Boolean |
| olddb_treatment | Character |
| openmpi_version | Character |
| organism | Character |
| other_seq | Boolean |
| other_sr_data | Boolean |
| outlier | Boolean |
| paper_name | Character |
| paper_nro_score | Float |
| paper_qc_score | Float |
| picard_date | Date |
| picard_dups_version | Character |
| picard_gc_version | Character |
| pipeline_hash | Character |
| pipeline_hash | Character |
| pipeline_revision_hash | Character |
| pipeline_revision_hash | Character |
| preseq_date | Date |
| Field name | Field type |
| preseq_version | Character |
| processing_notes | Character |
| protocol | Character |
| published | Boolean |
| python_version | Character |
| r_version | Character |
| raw_read_depth | Integer |
| raw_read_length | Integer |
| raw_strandedness | Character |
| rcomp | Boolean |
| replicate | Character |
| rna_seq | Boolean |
| rseqc_cds | Integer |
| rseqc_date | Date |
| rseqc_five_utr | Integer |
| rseqc_intron | Integer |
| rseqc_tags | Integer |
| rseqc_three_utr | Integer |
| rseqc_version | Character |
| rsubread_version | Character |
| sample_name | Character |
| sample_nro_score | Integer |
| sample_qc_score | Integer |
| sample_type | Character |
| samtools_version | Character |
| samtools_version | Character |
| search_field | Character |
| search_term | Character |
| seqkit_version | Character |
| single_map | Integer |
| single_paired | Character |
| spikein | Character |
| sratools_version | Character |
| srp | Character |
| srr | Character |
| start_time | Integer |
| strain | Character |
| tdf_avail | Boolean |
| tfit_avail | Boolean |
| tfit_bidir_gc | Float |
| tfit_date | Date |
| tfit_master_merge_incl | Boolean |
| tfit_prelim_date | Date |
| tfit_version | Character |
| three_dim_seq | Boolean |
| time_unit | Character |
| timecourse | Boolean |
| tissue | Character |
| tissue_description | Character |
| treatment | Character |
| trim_read_depth | Integer |
| unusable | Boolean |
| wildtype_untreated | Boolean |
| year | Integer |